|
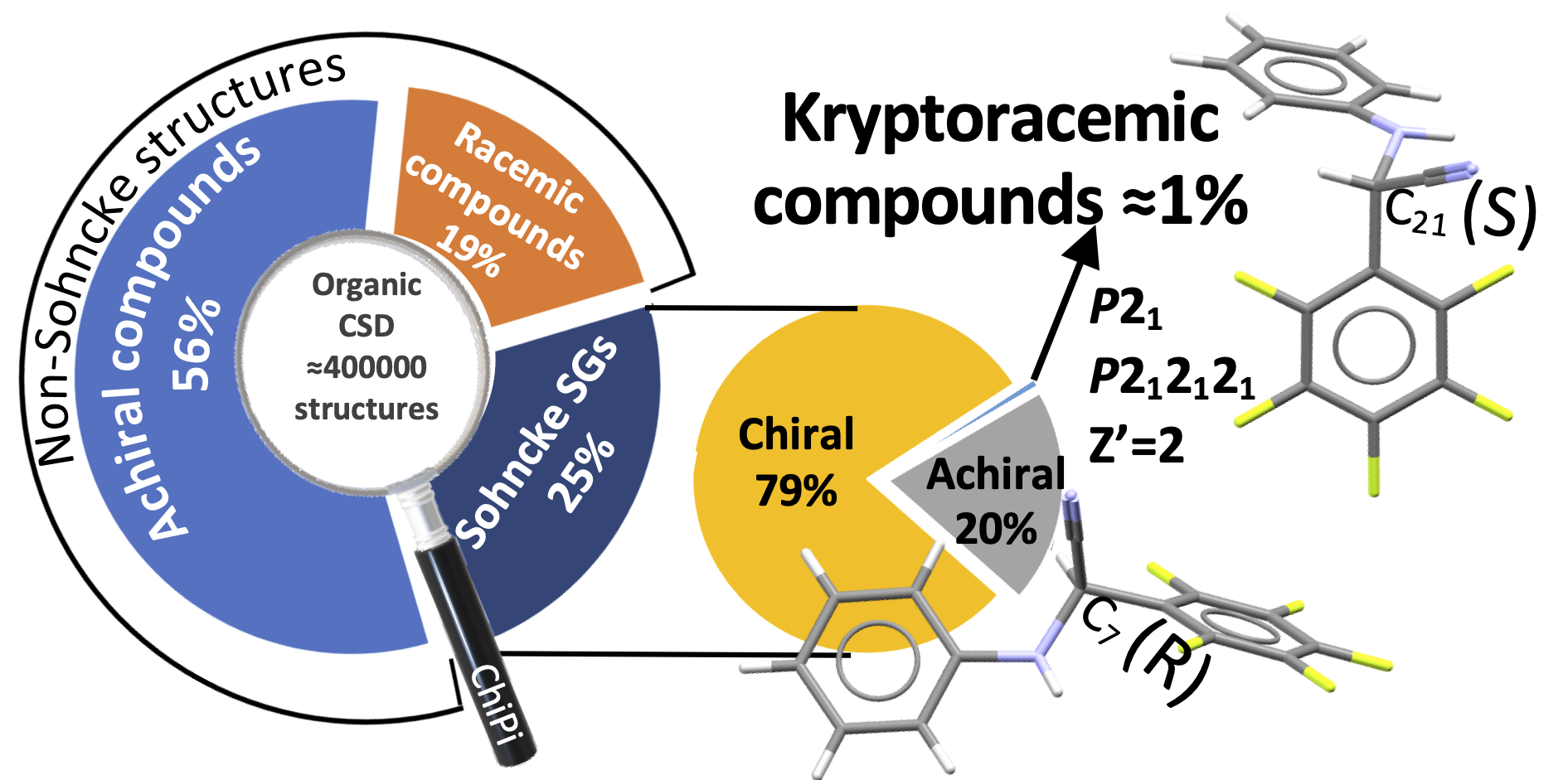
Disclaimer: the original code published in “Kryptoracemic Compound Hunting and Frequency in the Cambridge Structural Database,” CrystEngComm 43 (2020): 2286, doi:10.1039/D0CE00303D was written for Python 2 that is no longer supported. The new code available here is written in Python 3.7 now provided in new versions of CCDC. This version includes some improvements but results could differ slightly compare to the published version. Some new features are experimental and could not work correctly. |
About ChiPi
ChiPi is a python code that permits to check the molecular chirality in a given or a set of crystal structure(s) of the Cambridge Structural Database (CSD) and is based on the CCDC API.
RUN THE PROGRAM
Note: Before to start ChiPi you have to configure the CCDC Python API (see CCDC online installation notes: link)
To start ChiPi you need:
- The Chipi code (download link)
- A list of CCDC refcode you want to investigate (e.g. teaching_subset.gcd)
A refcode list can be generated through Conquest of the CCDC or manually using a text editor (Notepad for instance). Note that the last line should be a blank line to run ChiPi code.
How to run:
- Unzip Chipi.zip file in a working directory of your choice.
- Open a cmd (windows) or terminal (mac).
- Go to the folder containing the chipi source code. (> cd your_file_path)
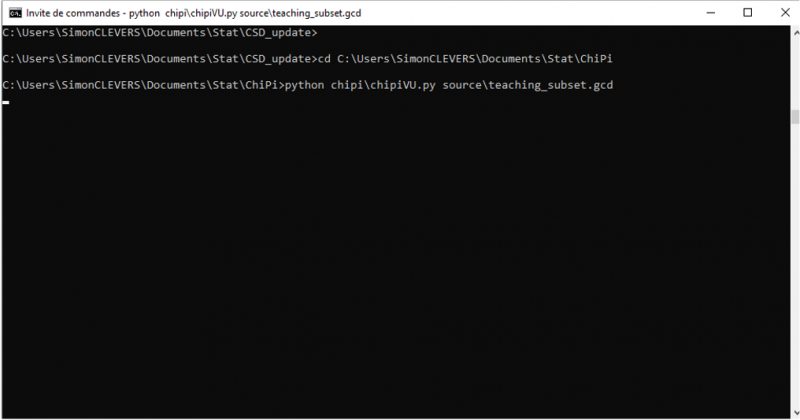
- Type the following command:
- “python chipi/chipyVU.py source/source.gcd”
Where “source.gcd” is the name of your refcode list (e.g. teaching_subset.gcd). The source.gcd file should be in the source folder.
!Warning! à Please check the validity of your refcodes before to start calculation.
If the warning "Not all the identifiers exist in the database" appears after the start of the code, it means that at least one CCDC Refcode is not in the CSD installed in your computer. The reasons could be:
- Refcode belonging to an old database version and cleaned since.
- An error in the refcode if you edit the “source.gcd” manually.
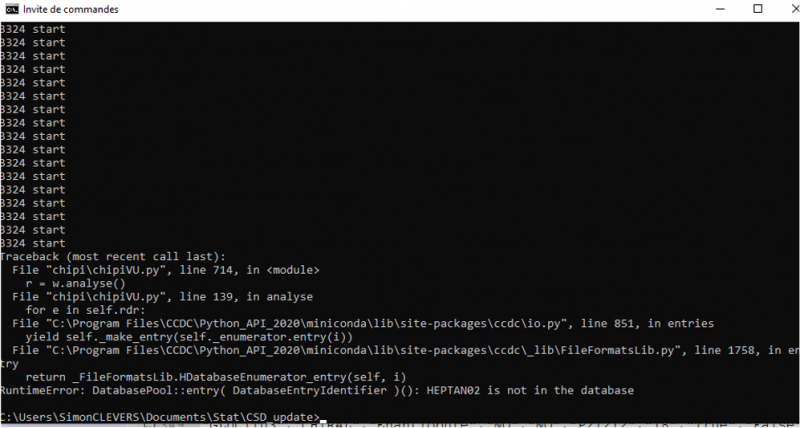
In this case, the program should quit with an error message when the wrong refcode is analyzed. Below example with the refcode HEPTAN02 that is no more in the database:
END of Calculation and Abort Command
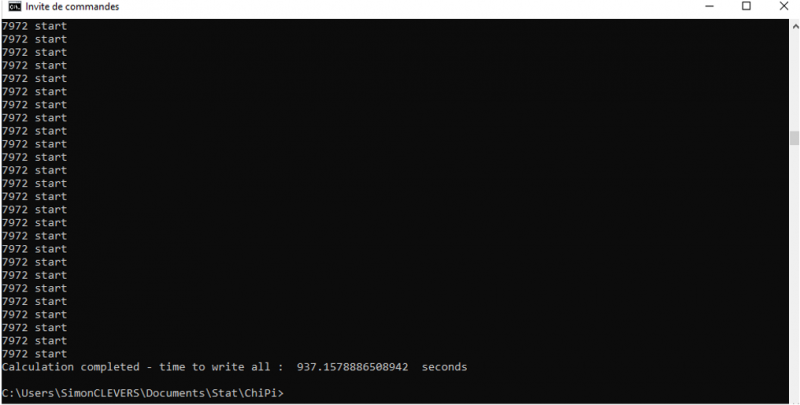
The end of the calculation is indicated by the message “Calculation completed – time to write all: X seconds.“ in the prompt. The result file is automatically generated in the “return” folder as “result.txt”
!Warning!: if you restart ChiPi, the previous result file will be erased and replaced! Copy your result files in a safe location.
This result file can then be opened by text editor or spreadsheet for postprocessing analyzes.
If for one reason you want to abort the calculation you can use the combination: ctrl+C in the terminal. Please not it could take time to abort if the program is in subroutine.
CONTACT
If an error occurs do not hesitate to contact: simon.clevers@gmail.com


